
{CopernicusMarine}Easily access information from https://data.marine.copernicus.eu
Copernicus Marine
Service Information is a programme subsidised by the European
Commission. Its mission is to provide free authoritative information on
the oceans physical and biogeochemical state. The
{CopernicusMarine} R package is developed apart from this
programme and facilitates retrieval of information from https://data.marine.copernicus.eu. With the package you
can:
{CopernicusMarine}Previously, there was only a MOTUS client available for Python.
Therefore, automating Copernicus downloads in R used to require
installation of python and its motuclient package. The
{CopernicusMarine} R package has a much simpler
installation procedure (see below) and does not depend on third party
software.
Get CRAN version
install.packages("CopernicusMarine")Get development version on github
devtools::install_github('pepijn-devries/CopernicusMarine')The package provides an interface between R and the Copernicus Marine services. Note that for some of these services you need an account and have to comply with specific terms. The usage section briefly shows three different ways of obtaining data from Copernicus:
Please check the manual for complete documentation of the package.
The code below assumes that you have registered your account details
using option(CopernicusMarine_uid = "my_user_name") and
option(CopernicusMarine_pwd = "my_password"). If you are
comfortable that it is secure enough, you can also store these options
in your .Rprofile such that they will be loaded each
session. Otherwise, you can also provide your account details as
arguments to the functions.
The example below demonstrates how to subset a specific layer for a
specific product. The subset is constrained by the region,
timerange, verticalrange and
sub_variables arguments. The subset is downloaded to the
temporary file specified with destination and can be read
using the {stars}
package.
destination <- tempfile("copernicus", fileext = ".nc")
copernicus_download_motu(
destination = destination,
product = "GLOBAL_ANALYSISFORECAST_PHY_001_024",
layer = "cmems_mod_glo_phy-cur_anfc_0.083deg_P1D-m",
variable = "sea_water_velocity",
output = "netcdf",
region = c(-1, 50, 10, 55),
timerange = c("2021-01-01", "2021-01-02"),
verticalrange = c(0, 2),
sub_variables = c("uo", "vo")
)
#> Logging in onto MOTU server...
#> Preparing download...
#> Downloading file...
#> Done
mydata <- stars::read_stars(destination)
#> vo, uo,
plot(mydata["vo"], col = hcl.colors(100), axes = T)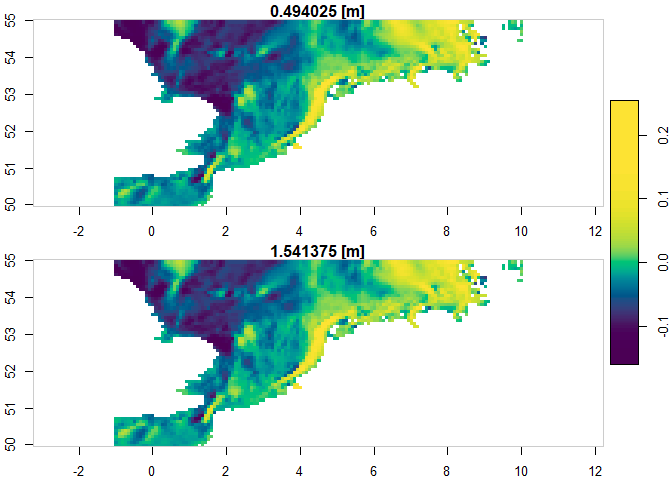
If you don’t want to subset the data and want the complete set, you can use the File Transfer Protocol (FTP) if that service is available for your product. First you can list files available for a specific product:
cop_ftp_files <- copernicus_ftp_list("GLOBAL_ANALYSISFORECAST_PHY_001_024", "cmems_mod_glo_phy-cur_anfc_0.083deg_P1D-m")
cop_ftp_files
#> # A tibble: 814 x 8
#> flags len protocol size month day time url
#> <chr> <int> <chr> <int> <chr> <int> <chr> <chr>
#> 1 -rw-rw-r-- 1 ftp 1937968861 Oct 14 06:10 ftp://nrt.cmems-du.eu~
#> 2 -rw-rw-r-- 1 ftp 1937978851 Oct 14 06:08 ftp://nrt.cmems-du.eu~
#> 3 -rw-rw-r-- 1 ftp 1937888467 Oct 14 06:07 ftp://nrt.cmems-du.eu~
#> 4 -rw-rw-r-- 1 ftp 1938380754 Oct 14 06:06 ftp://nrt.cmems-du.eu~
#> 5 -rw-rw-r-- 1 ftp 1938177028 Oct 14 06:09 ftp://nrt.cmems-du.eu~
#> 6 -rw-rw-r-- 1 ftp 1938234339 Oct 14 06:08 ftp://nrt.cmems-du.eu~
#> 7 -rw-rw-r-- 1 ftp 1938285902 Oct 14 06:08 ftp://nrt.cmems-du.eu~
#> 8 -rw-rw-r-- 1 ftp 1937954892 Oct 14 06:10 ftp://nrt.cmems-du.eu~
#> 9 -rw-rw-r-- 1 ftp 1937964478 Oct 14 06:09 ftp://nrt.cmems-du.eu~
#> 10 -rw-rw-r-- 1 ftp 1937962443 Oct 14 06:06 ftp://nrt.cmems-du.eu~
#> # ... with 804 more rowsDownloading the first file can be done with
copernicus_ftp_get(cop_ftp_files$url[[1]], tempdir()),
where the file would be stored in a temporary directory. By default the
progress is printed as files can be very large and may take some time to
download.
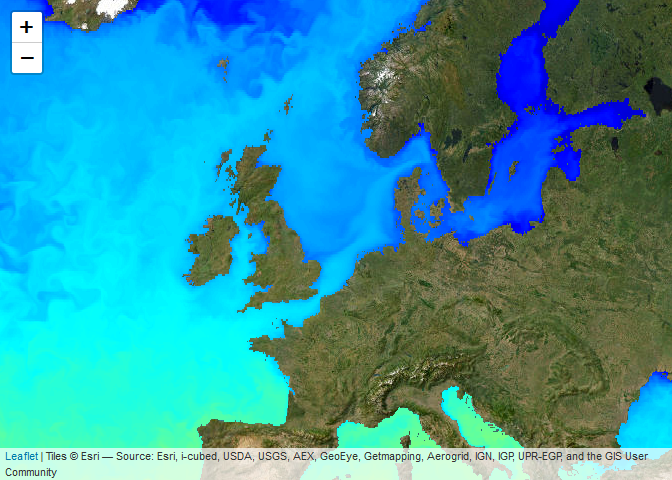
Web Map Services (WMS) allow to quickly plot pre-rendered images onto a map. This may not be useful when you need the data for analyses but is handy for quick visualisations, inspection or presentation of data. In R it is very easy to add WMS layers to an interactive map using leaflet widgets. This is demonstrated with the example below (note that in the documentation the map is only shown statically and is not interactive).
leaflet::leaflet() %>%
leaflet::setView(lng = 3, lat = 54, zoom = 4) %>%
leaflet::addProviderTiles("Esri.WorldImagery") %>%
addCopernicusWMSTiles(
product = "GLOBAL_ANALYSISFORECAST_PHY_001_024",
layer = "cmems_mod_glo_phy-thetao_anfc_0.083deg_P1D-m",
variable = "thetao"
)
When you want to use WMS tiles in static plots, you could download
and store specific regions of the tiles as a geo-referenced tiff
file for future use. Use copernicus_wms2geotiff for that
purpose.
A Copernicus account comes with several terms of use. One of these is that you properly cite the data you use in publications. In fact, we also have credit the data used in this documentation, which can be easily done with the following code:
copernicus_cite_product("GLOBAL_ANALYSISFORECAST_PHY_001_024")
#> $doi
#> [1] "Global Ocean 1/12° Physics Analysis and Forecast updated Daily - GLOBAL_ANALYSISFORECAST_PHY_001_024 (2016-10-14). DOI:10.48670/moi-00016"