

DriveML package for automated machine learning
especially in the classification context. DriveML saves a
lot of effort required for data preparation, feature engineering, model
selection and writing lengthy codes in a programming environment such as
R. Overall, the main benefits of DriveML are in development time
savings, reduce developers errors, optimal tuning of machine learning
models and reproducibility.
DriveML Framework:
DriveML is a series of functions such as
AutoDataPrep, AutoMAR,
autoMLmodel. DriveML automates some of the
most difficult machine learning functions such as data cleaning, data
transformations, feature engineering, model training, model validation,
model tuning and model selection.
Three key features of DriveML : Pre-processing, ML Techniques and Model interpretations
AutoDataPrep function to generate a novel features
based on the functional understanding of the datasetautoMLmodel function to develop baseline machine
learning models using regression and tree based classification
techniquesautoMLReport function to print the machine learning
model outcome in HTML formatInstall from CRAN within R using:
install.packages("DriveML")Install the latest development version of the DriveML from github with:
devtools::install_github("daya6489/DriveML")In this vignette, we will be using Heart Disease - Classifications data set
Data source UCI
library("DriveML")
library("SmartEDA")
## Load heart disease dataset
data(heart)
Understanding the dimensions of the dataset, variable names, overall missing summary and data types of each variables
## overview of the data;
ExpData(data = heart, type = 1)
## structure of the data
ExpData(data = heart, type = 2)To summarise the numeric variables, you can use following r codes from this pacakge
## Summary statistics by – overall
ExpNumStat(heart, by = "GA", gp = "target_var", Qnt = seq(0, 1, 0.1), MesofShape = 2, Outlier = TRUE, round = 2)## Generate Boxplot by category
ExpNumViz(heart, gp = "target_var", type = 2, nlim = 25, Page = c(2, 2))
## Generate Density plot
ExpNumViz(heart, gp = NULL, type = 3, nlim = 10, Page = c(2, 2))
## Generate Scatter plot
ExpNumViz(heart, target="target_var", nlim = 4, scatter = TRUE, Page=c(2, 1))One function to prepare a input data for machine learning model
# Data Preparation
small_data <- autoDataprep(heart, target = "target_var", missimpute = "default",
auto_mar = TRUE, mar_object = NULL, dummyvar = TRUE,
char_var_limit = 12, aucv = 0.02, corr = 0.99,
outlier_flag = TRUE, interaction_var = TRUE,
frequent_var = TRUE, uid = NULL, onlykeep = NULL, drop = NULL)
# Print output on R console
printautoDataprep(small_data)
# Final prepared master data
small_data_t <- small_data$master_dataOne function to develop machine learning binary classification model
# DriveML Model development
small_ml_random <- autoMLmodel(small_data_t, target = "target_var", testSplit = 0.2,
tuneIters = 5, tuneType = "random",
models = "all", varImp = 10, liftGroup = 10, maxObs = 10000, uid = NULL,
pdp = T, positive = 1, htmlreport = FALSE, seed = 1991)
# Model summary results
small_ml_random$resultsModel comparison results 
Test AUC 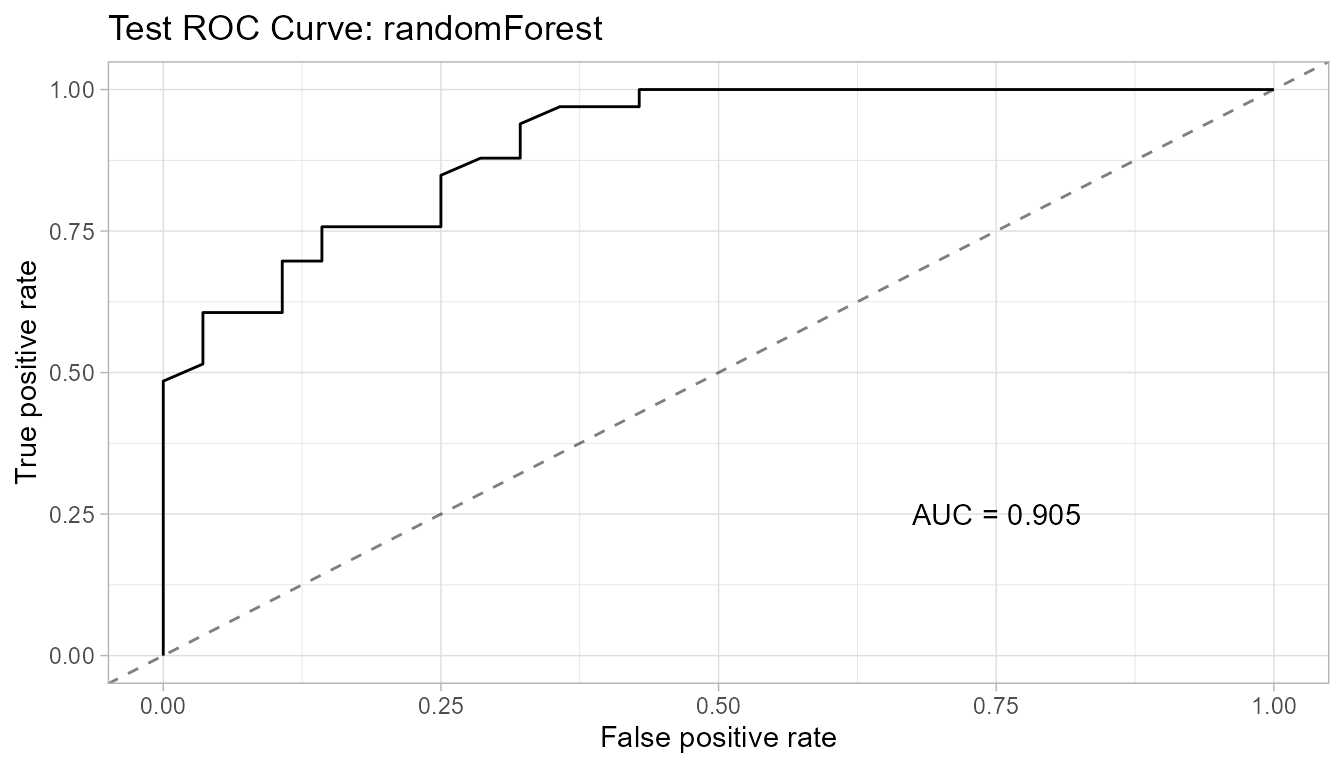
Variable Importance 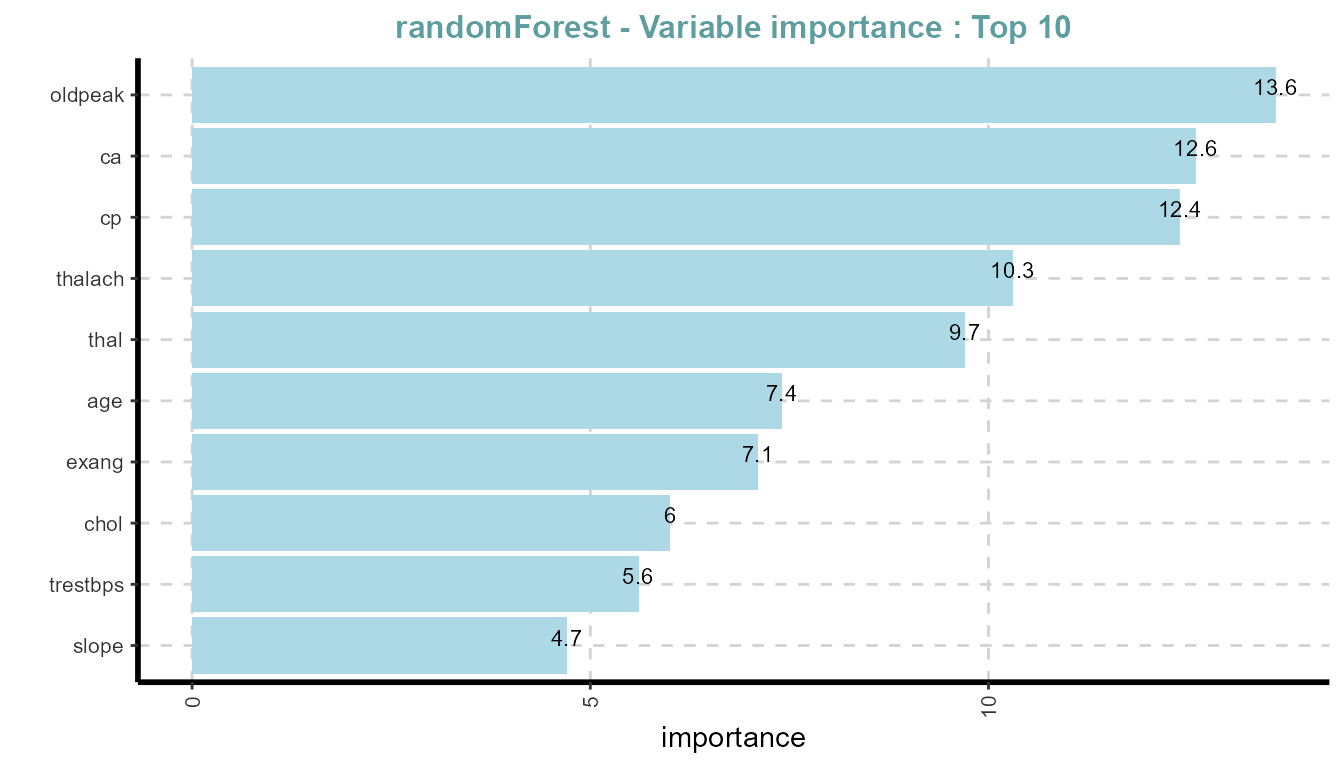
Threshold Plot 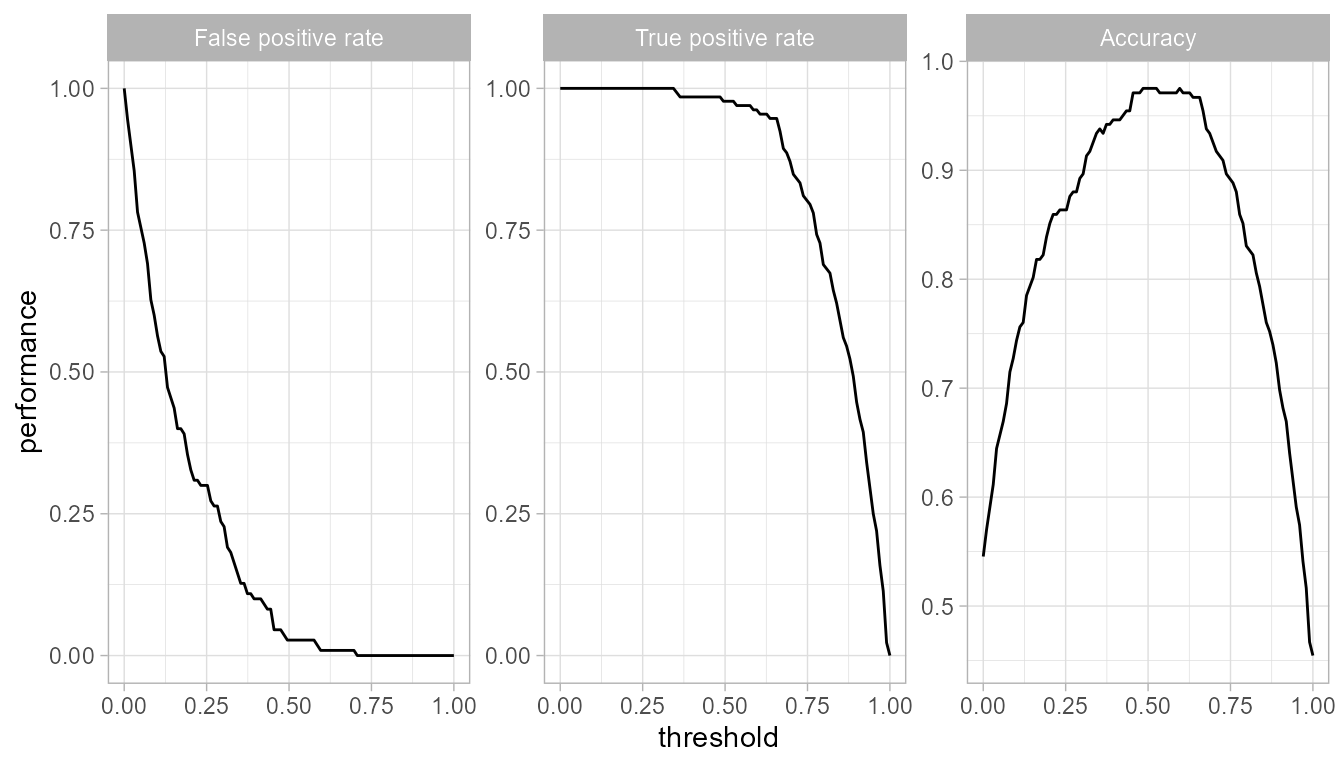
Generate a report in html format for the output of autoDataprep and autoMLmodel functions.
autoMLReport(mlobject = small_ml_random, mldata = small_data, op_file = "driveML_ouput_heart_data.html")The pre-print version of the paper on DriveML is available at ArXiv at- https://arxiv.org/pdf/2005.00478.pdf.
DriveML paper was presented at the 2nd International Workshop on Data Quality Assessment for Machine Learning at Knowledge Discovery and Data Mining (KDD) conference that was held on14-18 August, 2021, at Singapore. Conference workshop website link- http://data-readiness-kdd-2021.mybluemix.net
Boulange, A. (2020) automl: Deep Learning with Metaheuristic. URL:https://CRAN.R-project.org/package=automlr package version 1.3.2
Chen et al. (2020). xgboost: Extreme Gradient Boosting. URL:https://CRAN.R-project.org/package=xgboostr package version 1.0.0.2.
He, X., Zhao, K., & Chu, X. (2020). Automl: A survey of the state-of-the-art. arXiv:1908.00709v4. URL: https://arxiv.org/pdf/1908.00709.pdf
Therneau, T., & Atkinson, B. (2019). rpart: Recursive Partitioning and Regression Trees. URL: https://CRAN.R-project.org/package=rpart r package version 4.1-15.
Wright, M. N., & Ziegler, A. (2017). ranger: A fast implementation of random forests for high dimensional data in C ++ and R. Journal of Statistical Software, 77, 1–17. doi:10.18637/jss.v077.i01
Maher, M., & Sakr, S. (2019). Smartml: A meta learning-based framework for au-tomated selection and hyperparameter tuning for machine learning algorithms. In Advances in Database Technology-EDBT 2019: 22nd International Conference on Extending Database Technology . Lisbon, Portugal.