
The R package FFTrees creates, visualizes and evaluates fast-and-frugal decision trees (FFTs) for solving binary classification tasks following the methods described in Phillips, Neth, Woike & Gaissmaier (2017, as html | PDF).
Fast-and-frugal trees (FFTs) are simple and transparent decision algorithms for solving binary classification problems. The key feature making FFTs faster and more frugal than other decision trees is that every node allows for a decision. When predicting new outcomes, the performance of FFTs competes with more complex algorithms and machine learning techniques, such as logistic regression (LR), support-vector machines (SVM), and random forests (RF). Apart from being faster and requiring less information, FFTs tend to be robust against overfitting, and easy to interpret, use, and communicate.
The latest release of FFTrees is available from CRAN at https://CRAN.R-project.org/package=FFTrees:
install.packages("FFTrees")The current development version can be installed from its GitHub repository at https://github.com/ndphillips/FFTrees:
# install.packages("devtools")
devtools::install_github("ndphillips/FFTrees", build_vignettes = TRUE)As an example, let’s create a FFT predicting heart disease status
(Healthy vs. Diseased) based on the
heartdisease dataset included in
FFTrees:
library(FFTrees) # load packageThe heartdisease data provides medical information for
303 patients that were tested for heart disease. The full data were
split into two subsets: A heart.train dataset for fitting
decision trees, and heart.test dataset for a testing the
resulting trees. Here are the first rows and columns of both subsets of
the heartdisease data:
heart.train (the training / fitting dataset) contains
the data from 150 patients:head(heart.train)#> # A tibble: 6 × 14
#> diagnosis age sex cp trestbps chol fbs restecg thalach exang oldpeak
#> <lgl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 FALSE 44 0 np 108 141 0 normal 175 0 0.6
#> 2 FALSE 51 0 np 140 308 0 hypert… 142 0 1.5
#> 3 FALSE 52 1 np 138 223 0 normal 169 0 0
#> 4 TRUE 48 1 aa 110 229 0 normal 168 0 1
#> 5 FALSE 59 1 aa 140 221 0 normal 164 1 0
#> 6 FALSE 58 1 np 105 240 0 hypert… 154 1 0.6
#> # … with 3 more variables: slope <chr>, ca <dbl>, thal <chr>heart.test (the testing / prediction dataset) contains
data from a new set of 153 patients:head(heart.test)#> # A tibble: 6 × 14
#> diagnosis age sex cp trestbps chol fbs restecg thalach exang oldpeak
#> <lgl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 FALSE 51 0 np 120 295 0 hypert… 157 0 0.6
#> 2 TRUE 45 1 ta 110 264 0 normal 132 0 1.2
#> 3 TRUE 53 1 a 123 282 0 normal 95 1 2
#> 4 TRUE 45 1 a 142 309 0 hypert… 147 1 0
#> 5 FALSE 66 1 a 120 302 0 hypert… 151 0 0.4
#> 6 TRUE 48 1 a 130 256 1 hypert… 150 1 0
#> # … with 3 more variables: slope <chr>, ca <dbl>, thal <chr>Most of the variables in our data are potential predictors. The
(to-be predicted) criterion variable is diagnosis — a
logical column indicating the true state for each patient
(TRUE or FALSE, i.e., whether or not the
patient suffers from heart disease).
We use the main FFTrees() function to create FFTs for
the heart.train data and evaluate their predictive
performance on the heart.test data:
FFTrees object from the
heartdisease data:# Create an FFTrees object from the heartdisease data:
heart_fft <- FFTrees(formula = diagnosis ~.,
data = heart.train,
data.test = heart.test,
decision.labels = c("Healthy", "Disease"))FFTrees object shows basic information and
summary statistics (on the best training tree, FFT #1):# Print:
heart_fft#> FFTrees
#> - Trees: 7 fast-and-frugal trees predicting diagnosis
#> - Outcome costs: [hi = 0, mi = 1, fa = 1, cr = 0]
#>
#> FFT #1: Definition
#> [1] If thal = {rd,fd}, decide Disease.
#> [2] If cp != {a}, decide Healthy.
#> [3] If ca > 0, decide Disease, otherwise, decide Healthy.
#>
#> FFT #1: Training Accuracy
#> Training data: N = 150, Pos (+) = 66 (44%)
#>
#> | | True + | True - | Totals:
#> |----------|--------|--------|
#> | Decide + | hi 54 | fa 18 | 72
#> | Decide - | mi 12 | cr 66 | 78
#> |----------|--------|--------|
#> Totals: 66 84 N = 150
#>
#> acc = 80.0% ppv = 75.0% npv = 84.6%
#> bacc = 80.2% sens = 81.8% spec = 78.6%
#>
#> FFT #1: Training Speed, Frugality, and Cost
#> mcu = 1.74, pci = 0.87, E(cost) = 0.200FFTrees object (to visualize a tree and its performance) on
the test data:# Plot the best tree applied to the test data:
plot(heart_fft,
data = "test",
main = "Heart Disease")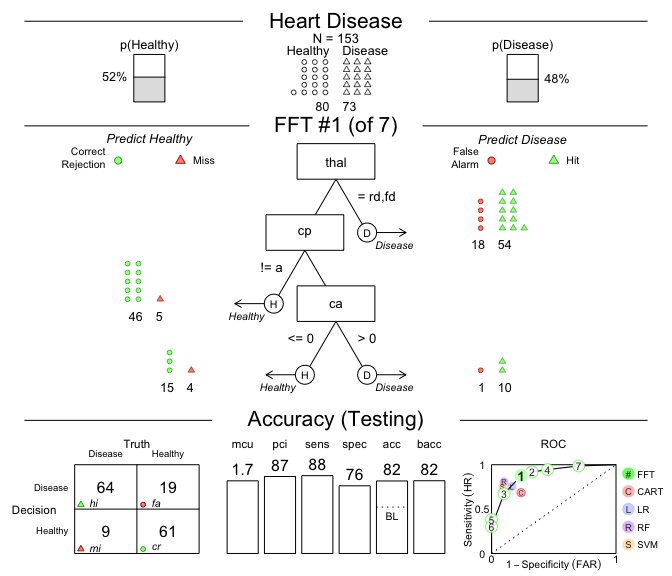
Figure 1: A fast-and-frugal tree (FFT) predicting
heart disease for test data and its performance
characteristics.
# Compare predictive performance across algorithms:
heart_fft$competition$test#> # A tibble: 5 × 17
#> algorithm n hi fa mi cr sens spec far ppv npv acc
#> <chr> <int> <int> <int> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 fftrees 153 64 19 9 61 0.877 0.762 0.238 0.771 0.871 0.817
#> 2 lr 153 55 13 18 67 0.753 0.838 0.162 0.809 0.788 0.797
#> 3 cart 153 50 19 23 61 0.685 0.762 0.238 0.725 0.726 0.725
#> 4 rf 153 59 8 14 72 0.808 0.9 0.1 0.881 0.837 0.856
#> 5 svm 153 55 7 18 73 0.753 0.912 0.0875 0.887 0.802 0.837
#> # … with 5 more variables: bacc <dbl>, wacc <dbl>, cost <dbl>, cost_dec <dbl>,
#> # cost_cue <dbl>FFTs are so simple that we even can create them ‘from words’ and then apply them to data!
For example, let’s create a tree with the following three nodes and
evaluate its performance on the heart.test data:
sex = 1, predict Disease.age < 45, predict Healthy.thal = {fd, normal}, predict Healthy,These conditions can directly be supplied to the my.tree
argument of FFTrees():
# Create custom FFT 'in words' and apply it to test data:
# 1. Create my own FFT (from verbal description):
my_fft <- FFTrees(formula = diagnosis ~.,
data = heart.train,
data.test = heart.test,
decision.labels = c("Healthy", "Disease"),
my.tree = "If sex = 1, predict Disease.
If age < 45, predict Healthy.
If thal = {fd, normal}, predict Healthy,
Otherwise, predict Disease.")
# 2. Plot and evaluate my custom FFT (for test data):
plot(my_fft,
data = "test",
main = "My custom FFT")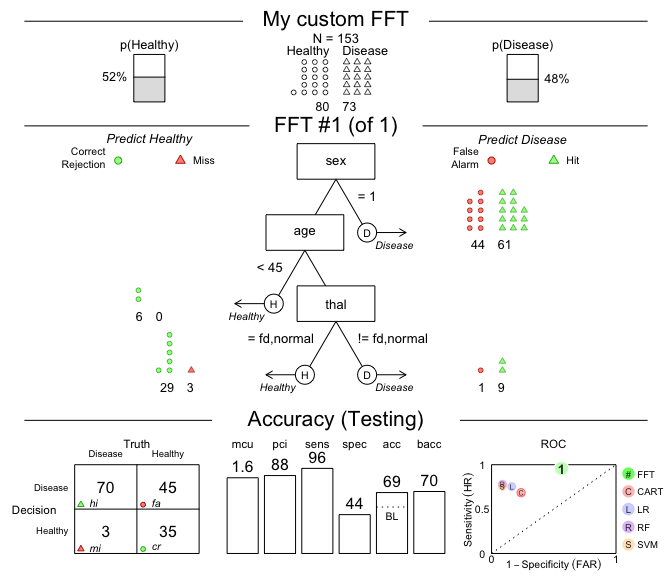
Figure 2: An FFT predicting heart disease created from a verbal description.
As we can see, this particular tree is somewhat biased: It has nearly
perfect sensitivity (i.e., is good at identifying cases of
Disease) but suffers from low specificity (i.e.,
performs poorly in identifying Healthy cases). Expressed in
terms of its errors, my_fft incurs few misses at the
expense of many false alarms. Although the accuracy of our
custom tree still exceeds the data’s baseline by a fair amount, the FFTs
in heart_fft (from above) strike a better balance.
Overall, what counts as the “best” tree for a particular problem depends on many factors (e.g., the goal of fitting vs. predicting data and the trade-offs between maximizing accuracy vs. incorporating the costs of cues or errors). To explore this range of options, the FFTrees package enables us to design and evaluate a range of FFTs.
We had a lot of fun creating FFTrees and hope you like it too! As a comprehensive, yet accessible introduction to FFTs, we recommend reading our article in the journal Judgment and Decision Making (2017, volume 12, issue 4), entitled FFTrees: A toolbox to create, visualize,and evaluate fast-and-frugal decision trees (available in html | PDF ).
Citation (in APA format):
We encourage you to read the article to learn more about the history of FFTs and how the FFTrees package creates, visualizes, and evaluates them. When using FFTrees in your own work, please cite us and share your experiences (e.g., on GitHub) so we can continue developing the package.
Here are some scientific publications that have used FFTrees (see Google Scholar for the full list):
[File README.Rmd last updated on 2023-01-06.]