deeptime extends the functionality of other plotting
packages like {ggplot2} and {lattice} to help
facilitate the plotting of data over long time intervals, including, but
not limited to, geological, evolutionary, and ecological data. The
primary goal of deeptime is to enable users to add
highly customizable timescales to their visualizations. Other functions
are also included to assist with other areas of deep time
visualization.
# get the stable version from CRAN
install.packages("deeptime")
# or get the development version from github
# install.packages("devtools")
devtools::install_github("willgearty/deeptime")library(deeptime)
library(tidyverse)The main function of deeptime is
coord_geo(), which functions just like
coord_trans() from {ggplot2}. You can use this
function to add highly customizable timescales to a wide variety of
ggplots.
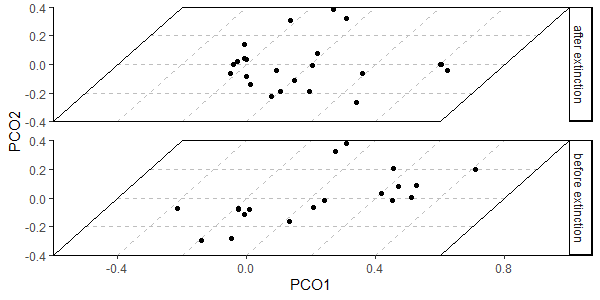
library(divDyn)
data(corals)
# this is not a proper diversity curve but it gets the point across
coral_div <- corals %>% filter(stage != "") %>%
group_by(stage) %>%
summarise(n = n()) %>%
mutate(stage_age = (stages$max_age[match(stage, stages$name)] +
stages$min_age[match(stage, stages$name)])/2)
ggplot(coral_div) +
geom_line(aes(x = stage_age, y = n)) +
scale_x_reverse("Age (Ma)") +
ylab("Coral Genera") +
coord_geo(xlim = c(250, 0), ylim = c(0, 1700)) +
theme_classic()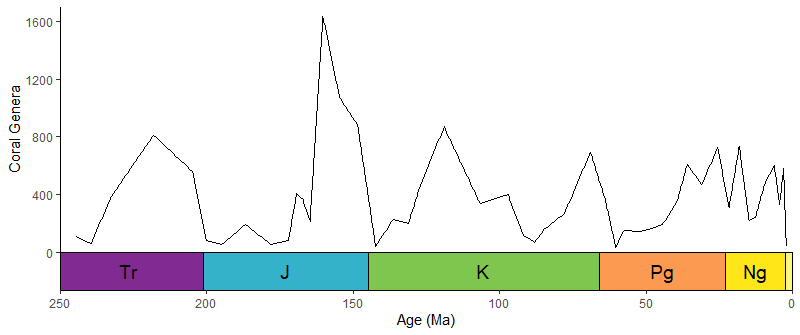
library(paleotree)
data(RaiaCopesRule)
p1 <- ggplot(ammoniteTraitsRaia) +
geom_point(aes(x = Log_D, y = FD)) +
labs(x = "Body size", y = "Suture complexity") +
theme_classic()
p2 <- ggplot(ammoniteTraitsRaia) +
geom_point(aes(x = Log_D, y = log_dur)) +
labs(x = "Body size", y = "Stratigraphic duration (myr)") +
theme_classic()
p3 <- ggtree(ammoniteTreeRaia, position = position_nudge(x = -ammoniteTreeRaia$root.time)) +
coord_geo(xlim = c(-415,-66), ylim = c(-2,Ntip(ammoniteTreeRaia)), pos = "bottom",
size = 4, abbrv = FALSE, neg = TRUE) +
scale_x_continuous(breaks = seq(-425, -50, 25), labels = -seq(-425, -50, 25)) +
theme_tree2() +
theme(plot.margin = margin(7,11,7,11))
ggarrange2(
ggarrange2(p1, p2, widths = c(2,1), draw = FALSE),
p3, nrow = 2, heights = c(1,2)
)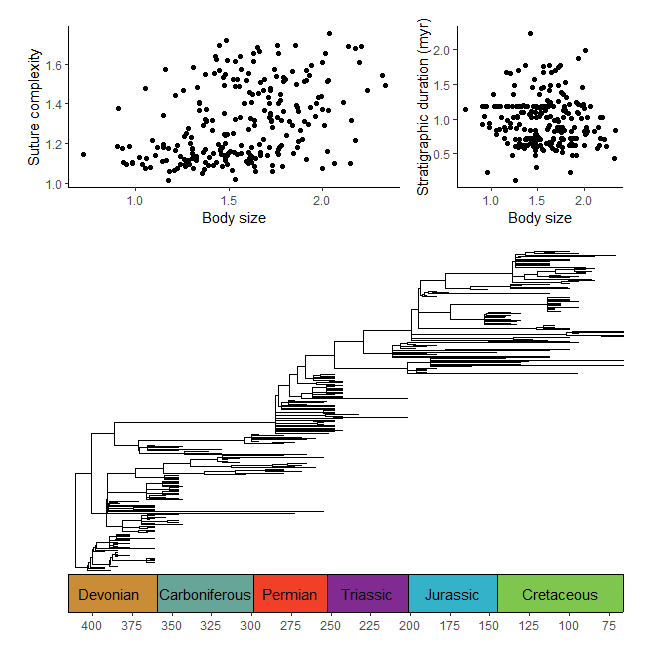
#make transformer
library(ggforce)
trans <- linear_trans(shear(.5, 0))
library(dispRity)
data(demo_data)
# prepare data to be plotted
crinoids <- as.data.frame(demo_data$wright$matrix[[1]][, 1:2])
crinoids$time <- "before extinction"
crinoids$time[demo_data$wright$subsets$after$elements] <- "after extinction"
square <- data.frame(V1 = c(-.6, -.6, .6, .6), V2 = c(-.4, .4, .4, -.4))
# plot data normally
ggplot() +
geom_segment(data = data.frame(x = -.6, y = seq(-.4, .4,.2),
xend = .6, yend = seq(-0.4, .4, .2)),
aes(x = x, y = y, xend = xend, yend=yend),
linetype = "dashed", color = "grey") +
geom_segment(data = data.frame(x = seq(-.6, .6, .2), y = -.4,
xend = seq(-.6, .6, .2), yend = .4),
aes(x = x, y = y, xend = xend, yend=yend),
linetype = "dashed", color = "grey") +
geom_polygon(data = square, aes(x = V1, y = V2), fill = NA, color = "black") +
geom_point(data = crinoids, aes(x = V1, y = V2), color = 'black') +
coord_cartesian(expand = FALSE) +
labs(x = "PCO1", y = "PCO2") +
theme_classic() +
facet_wrap(~time, ncol = 1, strip.position = "right") +
theme(panel.spacing = unit(1, "lines"), panel.background = element_blank())
# plot data with transformation
ggplot() +
geom_segment(data = data.frame(x = -.6, y = seq(-.4, .4,.2),
xend = .6, yend = seq(-0.4, .4, .2)),
aes(x = x, y = y, xend = xend, yend=yend),
linetype = "dashed", color = "grey") +
geom_segment(data = data.frame(x = seq(-.6, .6, .2), y = -.4,
xend = seq(-.6, .6, .2), yend = .4),
aes(x = x, y = y, xend = xend, yend=yend),
linetype = "dashed", color = "grey") +
geom_polygon(data = square, aes(x = V1, y = V2), fill = NA, color = "black") +
geom_point(data = crinoids, aes(x = V1, y = V2), color = 'black') +
coord_trans_xy(trans = trans, expand = FALSE) +
labs(x = "PCO1", y = "PCO2") +
theme_classic() +
facet_wrap(~time, ncol = 1, strip.position = "right") +
theme(panel.spacing = unit(1, "lines"), panel.background = element_blank())