This package can be used to easily access the data of the EBV netCDFs which can be downloaded from the EBV Data Portal. It also provides some basic visualization. Advanced users can build their own netCDFs following the EBV structure.
The EBV netCDF structure is designed to hold Essential Biodiversity Variables. This concept is further described here. The files are based on the Network Common Data Format (netCDF). Additionally, it follows the Climate and Forecast Conventions (CF, version 1.8) and the Attribute Convention for Data Discovery (ACDD, version 1.3).
The structure allows several datacubes per netCDF file. These cubes have four dimensions: longitude, latitude, time and entity, whereby the last dimension can, e.g., encompass different species or groups of species, ecosystem types or other. The usage of hierarchical groups enables the coexistence of multiple EBV cubes. The first level (netCDF group) are scenarios, e.g., the modelling for different Shared Socioeconomic Pathways (SSP) scenarios. The second level (netCDF group) are metrics, e.g., the percentage of protected area per pixel and its proportional loss over a certain time span per pixel. All metrics are repeated per scenario, if any are present.
├── scenario_1
│ └── metric_1
│ └── ebv_cube [lon, lat, time, entity]
└── scenario_2
└── metric_2
└── ebv_cube [lon, lat, time, entity]Just keep in mind: All EBV netCDF always have a metric. But they may or may not have a scenario. The resulting datacubes hold the data. These datacubes are 4D.
You can install the ebvcube packages with:
devtools::install_github('https://github.com/LuiseQuoss/ebvcube')This packages uses GDAL tools (GDAL version: 3.1.4). You need a GDAL installation on your machine. One possibility to install GDAL is the OSGeo4W Network installer. Check GDAL when running the installation! If you have QGIS on your machine, GDAL should be included. If you have problems you can set the GDAL related paths by hand using the following lines of code. Your paths will differ! First check your GDAL installation.
#add GDAL path to the existing paths
Sys.setenv(PATH = paste0('C:\\OSGeo4W64\\bin;',Sys.getenv("PATH")))
#check and change path for proj_lib, gdal_data and gdal_driver_path
Sys.setenv(PROJ_LIB = 'C:\\OSGeo4W64\\share\\proj')
Sys.setenv(GDAL_DATA = 'C:\\OSGeo4W64\\share\\gdal')
Sys.setenv(GDAL_DRIVER_PATH = 'C:\\OSGeo4W64\\bin\\gdalplugins')
#you can always check your GDAL path settings using
Sys.getenv("PATH")
Sys.getenv("PROJ_LIB")
Sys.getenv("GDAL_DATA")
Sys.getenv("GDAL_DRIVER_PATH")With the following two functions you get the core information about the data of a specific EBV netCDF. First we take a look at some basic metadata of that file. The properties encompass much more information!
library(ebvcube)
#set the path to the file
file <- system.file(file.path("extdata","martins_comcom_id1_20220208_v1.nc"), package="ebvcube")
#read the properties of the file
prop.file <- ebv_properties(file)
#take a look at the general properties of the dataset - there are more properties to discover!
prop.file@general[1:4]
#> $title
#> [1] "Local bird diversity (cSAR/BES-SIM)"
#>
#> $description
#> [1] "Changes in bird diversity at 1-degree resolution caused by land use, estimated by the cSAR model for 1900-2015 using LUH2.0 historical reconstruction of land-use."
#>
#> $ebv_class
#> [1] "Community composition"
#>
#> $ebv_name
#> [1] "Taxonomic and phylogenetic diversity"
slotNames(prop.file)
#> [1] "general" "spatial" "temporal" "metric" "scenario" "ebv_cube"Now let’s get the paths to all possible datacubes. The resulting dataframe includes the paths and also descriptions of the metric and/or scenario and/or entity. The paths basically consist of the nested structure of scenario, metric and the datacube.
datacubes <- ebv_datacubepaths(file, verbose=FALSE)
datacubes
#> datacubepaths metric_names
#> 1 metric_1/ebv_cube Relative change in the number of species (%)
#> 2 metric_2/ebv_cube Absolute change in the number of speciesIn the next step we will get the properties of one specific datacube - fyi: the result also holds the general file properties from above.
prop.dc <- ebv_properties(file, datacubes[1,1])
prop.dc@ebv_cube
#> $units
#> [1] "Percentage points"
#>
#> $coverage_content_type
#> [1] "modelResult"
#>
#> $fillvalue
#> [1] -3.4e+38
#>
#> $type
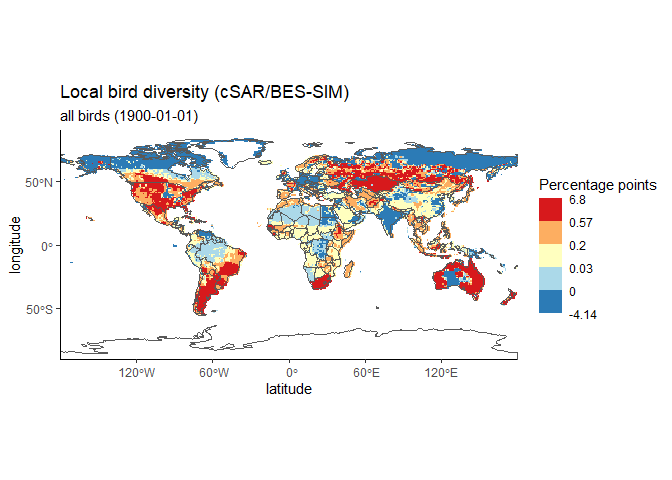
#> [1] "H5T_IEEE_F32LE"To discover the spatial distribution of the data, you can plot a map of the datacube that we just looked at. It has 12 timesteps. Here we look at the sixth one.
#plot the global map
dc <- datacubes[1,1]
ebv_map(file, dc, entity=1, timestep = 1)
# What was the data about again? Check the properties!
prop.dc@general$title
#> [1] "Local bird diversity (cSAR/BES-SIM)"
# And the datacube?
prop.dc@metric$name
#> [1] "Relative change in the number of species (%)"
prop.dc@general$entity_names[1]
#> [1] "all birds"
#What timestep?
prop.dc@temporal$timesteps_natural[1]
#> [1] "1900-01-01"It’s nice to see the global distribution, but how is the change of that datacube (non forest birds) over time? Let’s take a look at the average. The function returns the values, catch them!
#get the averages and plot
averages <- ebv_trend(file, dc, entity=1)
#> [1] "calculating timesteps..."
#> ================================================================================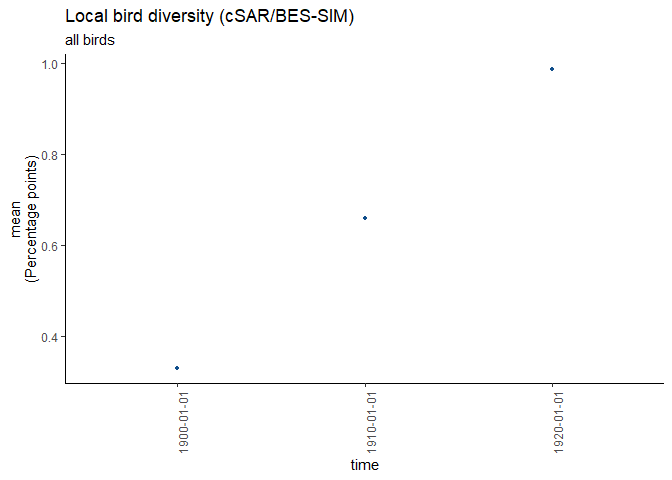
averages
#> [1] 0.3302859 0.6599264 0.9860116It would be cool to have that for other indicators as well? Check out the different options for ‘method’.
Before you actually load the data it may be nice to get an impression of the value range and other basic measurements.
#info for whole dataset
measurements <- ebv_analyse(file, dc, entity=1)
#see the included measurements
names(measurements)
#> [1] "min" "q25" "q50" "mean" "q75" "max" "std" "n" "NAs"
#how many pixels are included?
measurements$n
#> [1] 64800
measurements$mean
#> [1] 0.3302859
#info for a subset defined by a bounding box (roughly(!) Germany)
bb <- c(5,15,47,55)
measurements.bb <- ebv_analyse(file, dc, entity = 1, subset = bb)
#how many pixels are now included?
measurements.bb$n
#> [1] 80
measurements.bb$mean
#> [1] -0.8185277To access the data you can use the following:
#load whole data as array for two timesteps
data <- ebv_read(file, dc, entity = 1, timestep = c(1,2), type = 'a')
dim(data)
#> [1] 180 360 2To subset the data using a shapefile you need to indicate a directory for temporarily created files.
#load subset from shapefile (Germany)
shp <- system.file(file.path('extdata','subset_germany.shp'), package="ebvcube")
data.shp <- ebv_read_shp(file, dc, entity=1, shp = shp, timestep = c(1,2,3))
dim(data.shp)
#> [1] 9 11 3
#very quick plot of the resulting raster plus the shapefile
borders <- terra::vect(shp)
ggplot2::ggplot() +
tidyterra::geom_spatraster(data = data.shp[[1]]) +
tidyterra::geom_spatvector(data = borders, fill = NA) +
ggplot2::scale_fill_fermenter(na.value=NA, palette = 'YlGn', direction = 1) +
ggplot2::theme_classic()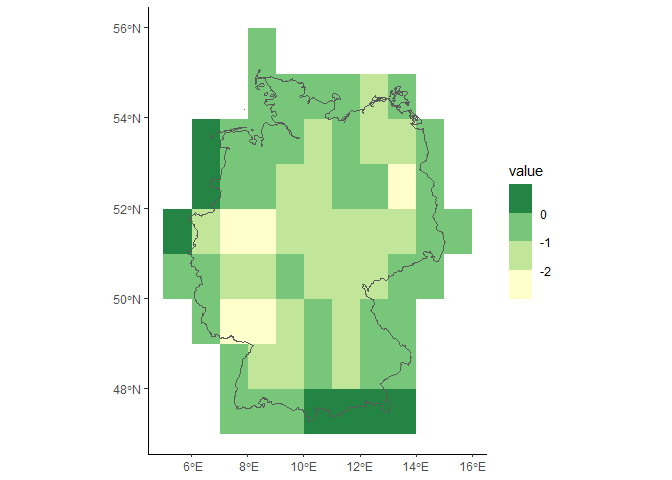 Imagine you have a very large dataset but only limited memory. The
package provides the possibility to load the data as a DelayedArray. A
second function helps you to write that data back on disk properly. Look
into the manual to obtain more information.
Imagine you have a very large dataset but only limited memory. The
package provides the possibility to load the data as a DelayedArray. A
second function helps you to write that data back on disk properly. Look
into the manual to obtain more information.
First of all, you have to insert all the metadata in the EBV Data Portal and then use the resulting text file (json format) to create an empty netCDF which complies to the EBV netCDF structure, i.e., it has the correct structure mapped to your data and holds the metadata. Additionally to that (json) text file the function needs a list of all entities the netCDF (csv list, see help page for detailed information) will encompass and geospatial information such as the coordinate reference system.
The example is based on the Local bird diversity (cSAR/BES-SIM).
#paths
json <- system.file(file.path('extdata','metadata.json'), package="ebvcube")
newNc <- file.path(system.file(package="ebvcube"),'extdata','test.nc')
entities <- file.path(system.file(package='ebvcube'),"extdata","entities.csv")
#defining the fillvalue - optional
fv <- -3.4e+38
#create the netCDF
ebv_create(jsonpath = json, outputpath = newNc, entities = entities,
epsg = 4326, extent = c(-180, 180, -90, 90), resolution = c(1, 1),
fillvalue = fv, prec='float', force_4D = TRUE, overwrite=T, verbose=FALSE)
#needless to say: check the properties of your newly created file to see if you get what you want
#especially the entity_names from the slot general should be checked to see if your csv was formatted the right way
print(ebv_properties(newNc)@general$entity_names)
#> [1] "forest bird species" "non-forest bird species"
#> [3] "all bird species"
#check out the (still empty) datacubes that are available
dc.new <- ebv_datacubepaths(newNc, verbose=FALSE)
print(dc.new)
#> datacubepaths metric_names
#> 1 metric_1/ebv_cube Relative change in the number of species (%)
#> 2 metric_2/ebv_cube Absolute change in the number of speciesHint: You can always take a look at your netCDF in Panoply provided by NASA. That’s very helpful to understand the structure.
In the next step you can add your data to the netCDF from GeoTiff files or in-memory objects (matrix/array). You need to indicate the datacubepath the data belongs to. You can add your data timestep per timestep, in slices or all at once. You can simply add more data to the same datacube by changing the timestep definition.
#path to tif with data
root <- system.file(file.path('extdata'), package="ebvcube")
tifs <- c('entity1.tif', 'entity2.tif', 'entity3.tif')
tif_paths <- file.path(root, tifs)
#adding the data
entity <- 1
for (tif in tif_paths){
ebv_add_data(filepath_nc = newNc, datacubepath=dc.new[1,1], entity = entity,
timestep=1:3, data = tif, band=1:3)
entity <- entity + 1
}Ups! So you did a mistake and want to change the attribute?! No problem. Just use the upcoming function to change it.
ebv_attribute(newNc, attribute_name='units', value='percentage', levelpath=dc.new[1,1])
#check the properties one more time - perfect!
print(ebv_properties(newNc, dc.new[1,1])@ebv_cube$units)
#> [1] "percentage"In this case the levelpath corresponds to the datacube path. But you can also alter attributes at the metric or scenario level. See the manual for more info.
citation('ebvcube')
#>
#> To cite ebvcube in publications use:
#>
#> Quoss L, Fernandez N, Langer C, Valdez J, Pereira H (2021). _ebvcube:
#> Working with netCDF for Essential Biodiversity Variables_. German
#> Centre for Integrative Biodiversity Research (iDiv)
#> Halle-Jena-Leipzig, Germany. R package version 0.1.1,
#> <https://github.com/LuiseQuoss/ebvcube>.
#>
#> Ein BibTeX-Eintrag für LaTeX-Benutzer ist
#>
#> @Manual{,
#> title = {ebvcube: Working with netCDF for Essential Biodiversity Variables},
#> author = {Luise Quoss and Nestor Fernandez and Christian Langer and Jose Valdez and Henrique Miguel Pereira},
#> year = {2021},
#> note = {R package version 0.1.1},
#> organization = {German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig},
#> address = {Germany},
#> url = {https://github.com/LuiseQuoss/ebvcube},
#> }| Functionality | Function | Description |
|---|---|---|
| Basic access | ebv_datacubepaths | Returns all available data cubes in the netCDF |
| ebv_properties | Collects all the metadata terms of the netCDF | |
| ebv_download | Downloads EBV netCDFs from the EBV Portal | |
| Data access | ebv_read | Reads the data |
| ebv_read_bb | Reads a spatial subset given by a bounding box | |
| ebv_read_shp | Reads a spatial subset given by a Shapefile | |
| ebv_analyse | Returns basic measurements of the data | |
| ebv_write | Writes manipulated data back to disc | |
| Data visualization | ebv_map | Plots a map of the specified data slice |
| ebv_trend | Plots the temporal trend | |
| Data creation | ebv_create | Creates a new EBV netCDF |
| ebv_add_data | Adds data to the new netCDF | |
| ebv_attribute | Changes attribute values |