
exDE provides tools to set up modular ordinary and delay differential
equation spatial models for mosquito-borne pathogens, focusing on
malaria. Modularity is achieved by method dispatch on parameter lists
for each component which is used to compute the full set of differential
equations. The function exDE::xDE_diffeqn computes the
gradient of all state variables from those modular components and can be
used with the excellent solvers in deSolve, or other
differential equation solvers in R. exDE can be regarded as the
continuous-time companion to the discrete stochastic Micro-MoB framework.
To get started, please consider reading some of the articles in the drop down panels above, at our website. The 3 sections ending in “Component” describe particular models implementing the interface for each of those components (adult mosquitoes, aquatic mosquitoes, and humans), and show a simulation at their equilibrium values.
The section “Articles” has more in-depth examples, including an
extended walk through of how to set up and run a model in
vignette("ex_534"), a guide on how to contribute, and an
example of running a model in exDE with external forcing under a model
of ITN (insecticide treated nets) based vector control in
vignette("vc_lemenach").
The section “Functions” documents each function exported by the package.
To install from an R session, run the following lines of code.
library(devtools)
devtools::install_github("dd-harp/exDE")For information about how to contribute to the development of exDE,
please read our article on how to contribute at
vignette("Contributing")!
Models for mosquito borne pathogen transmission systems are naturally modular, structured by vector life stage, host population strata, and by the spatial locations (patches) at which transmission occurs (see figure below).
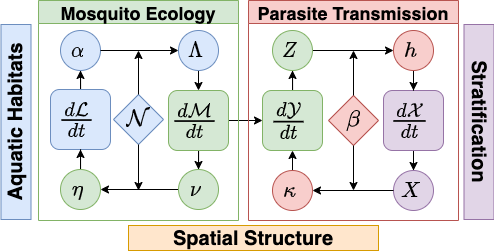
Models in the exDE framework are constructed from 3 dynamical components:
The combined state from these 3 components is the entire state of the dynamical model, and their combined dynamics described by their differential equations represents the full endogenous dynamics of the system. In addition there are 2 more components which do not directly contribute to the state of the model, but instead modify parameters and compute intermediate quantities to represent external influences on the system. These are:
There are also functions which handle the exchange of information (flows) between the dynamical components and which couple their dynamics. Bloodfeeding is the process by which adult mosquitoes seek out and feed on blood hosts, and results in the quantities \(EIR\) (entomological inoculation rate) and \(\kappa\), the net infectiousness of humans to mosquitoes, which couple the dynamics of \(\mathcal{M}\) and \(\mathcal{X}\). Likewise emergence of new adults from aquatic habitats and egg laying by adults into habitats couples \(\mathcal{M}\) and \(\mathcal{L}\).
The function exDE::xDE_diffeqn compute the necessary
quantities and returns a vector of derivatives of all state variables
which can be used to solve trajectories from a model in exDE. The
program flow within this function is summarized by this diagram:
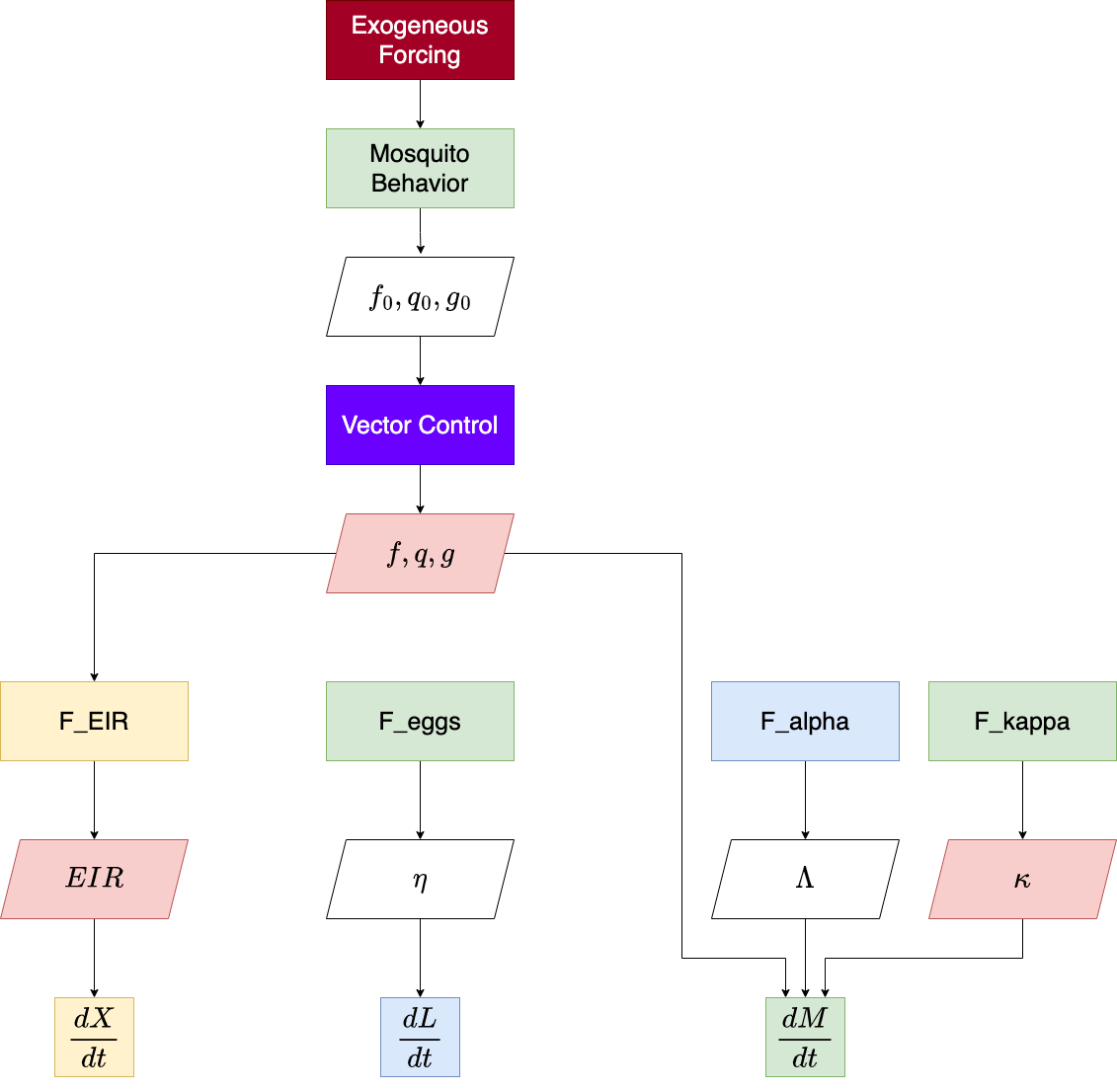
For more information, please read our research article describing the theory behind the model.