gips
gips
gips - Gaussian model Invariant by Permutation Symmetry
gips is an R package that looks for permutation
symmetries in the multivariate Gaussian sample. Such symmetries reduce
the free parameters in the unknown covariance matrix. This is especially
useful when the number of variables is substantially larger than the
number of observations.
gips will help
you with two things:gips
can be used as an exploratory tool for searching the space of
permutation symmetries of the Gaussian vector. Useful in the Exploratory
Data Analysis (EDA).From CRAN:
# Install the released version from CRAN:
install.packages("gips")From GitHub:
# Install the development version from GitHub:
# install.packages("devtools")
devtools::install_github("PrzeChoj/gips")Assume we have the data, and we want to understand its structure:
library(gips)
Z <- HSAUR::aspirin
# Renumber the columns for better readability:
Z[,c(2,3)] <- Z[,c(3,2)]Assume the data Z is normally distributed.
dim(Z)
#> [1] 7 4
number_of_observations <- nrow(Z) # 7
perm_size <- ncol(Z) # 4
S <- cov(Z)
round(S)
#> [,1] [,2] [,3] [,4]
#> [1,] 381405 345527 1864563 1813725
#> [2,] 345527 316411 1711853 1663065
#> [3,] 1864563 1711853 9305049 8991343
#> [4,] 1813725 1663065 8991343 8755176
g <- gips(S, number_of_observations)
my_add_text(plot(g, type = "heatmap"))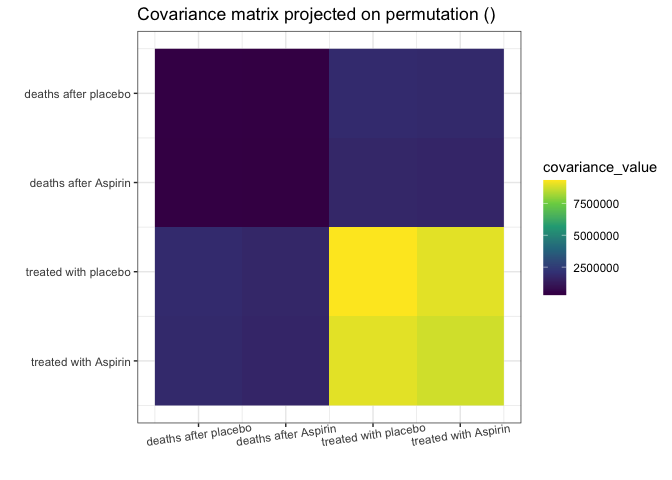
Remember, we analyze the covariance matrix. We can see some strong
similarities between the covariances of columns 3 and 4. Those have
similar variances (S[3,3]
S[4,4]), and their covariances
with the rest of the columns are alike (S[1,3]
S[1,4] and S[2,3]
S[2,4]).
Note that the variances of columns 1 and 2 are also similar, but the covariances with other columns (3 and 4) are not alike.
Let’s see if the find_MAP() will find this
relationship:
g_MAP <- find_MAP(g, optimizer = "brute_force")
#> ================================================================================
g_MAP
#> The permutation (3,4)
#> - was found after 24 log_posteriori calculations
#> - is 106222567640989 times more likely than the starting, () permutation.The find_MAP found the relationship (3,4). In its
opinion, the variances [3,3] and [4,4] are so close to each other that
it is reasonable to consider them equal. Similarly, the covariances
[1,3] and [1,4]; just as covariances [2,3] and [3,4], also will be
considered equal:
S_projected <- project_matrix(S, g_MAP[[1]])
round(S_projected)
#> [,1] [,2] [,3] [,4]
#> [1,] 381405 345527 1839144 1839144
#> [2,] 345527 316411 1687459 1687459
#> [3,] 1839144 1687459 9030113 8991343
#> [4,] 1839144 1687459 8991343 9030113
my_add_text(plot(g_MAP, type = "heatmap"))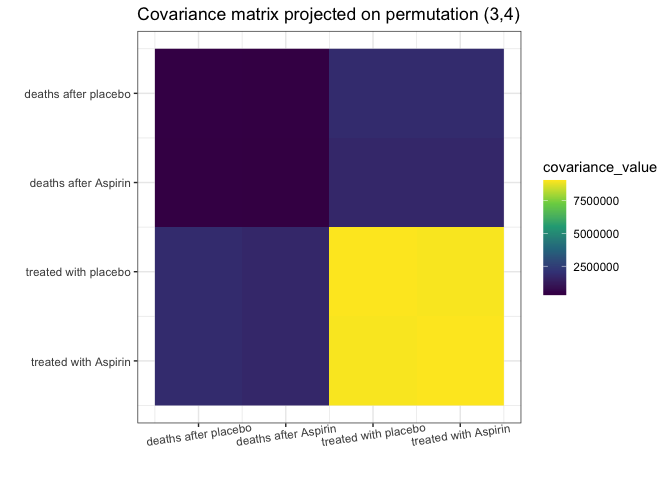
This S_projected matrix can now be interpreted as a more
stable covariance matrix estimator.
We can also interpret the data suggesting there is, for example, the same covariance of “number of deaths after Aspirin” with “number of people treated with *” no matter if the “*” represents the placebo or Aspirin.
First, construct data for the example:
# Prepare model, multivariate normal distribution
perm_size <- 6
mu <- numeric(perm_size)
sigma_matrix <- matrix(
data = c(
1.1, 0.8, 0.6, 0.4, 0.6, 0.8,
0.8, 1.1, 0.8, 0.6, 0.4, 0.6,
0.6, 0.8, 1.1, 0.8, 0.6, 0.4,
0.4, 0.6, 0.8, 1.1, 0.8, 0.6,
0.6, 0.4, 0.6, 0.8, 1.1, 0.8,
0.8, 0.6, 0.4, 0.6, 0.8, 1.1
),
nrow = perm_size, byrow = TRUE
) # sigma_matrix is a matrix invariant under permutation (1,2,3,4,5,6)
# Generate example data from a model:
Z <- MASS::mvrnorm(4, mu = mu, Sigma = sigma_matrix)
# End of prepare modelSuppose we do not know the true covariance matrix and we want to estimate it. We cannot use the
standard MLE because it does not exists (
,
).
We will assume it was generated from the normal distribution with the
mean .
library(gips)
dim(Z)
#> [1] 4 6
number_of_observations <- nrow(Z) # 4
perm_size <- ncol(Z) # 6
# Calculate the covariance matrix from the data:
S <- (t(Z) %*% Z) / number_of_observationsMake the gips object out of data:
g <- gips(S, number_of_observations, was_mean_estimated = FALSE)Find the Maximum A Posteriori Estimator for the permutation. Space is
small (), so it is reasonable to browse the
whole of it:
g_map <- find_MAP(g, optimizer = "brute_force")
#> ================================================================================
g_map
#> The permutation (1,2,3,4,5,6)
#> - was found after 720 log_posteriori calculations
#> - is 234.659014400441 times more likely than the starting, () permutation.We see that the found permutation is hundreds of times more likely than making no additional assumption. That means the additional assumptions are justified.
summary(g_map)$n0
#> [1] 1
summary(g_map)$n0 <= number_of_observations # 1 <= 4
#> [1] TRUEWhat is more, we see the number of observations () is bigger or equal to
, so we can estimate the covariance
matrix with the Maximum Likelihood estimator:
S_projected <- project_matrix(S, g_map[[1]])
S_projected
#> [,1] [,2] [,3] [,4] [,5] [,6]
#> [1,] 1.0432544 0.8066746 0.5881748 0.4007521 0.5881748 0.8066746
#> [2,] 0.8066746 1.0432544 0.8066746 0.5881748 0.4007521 0.5881748
#> [3,] 0.5881748 0.8066746 1.0432544 0.8066746 0.5881748 0.4007521
#> [4,] 0.4007521 0.5881748 0.8066746 1.0432544 0.8066746 0.5881748
#> [5,] 0.5881748 0.4007521 0.5881748 0.8066746 1.0432544 0.8066746
#> [6,] 0.8066746 0.5881748 0.4007521 0.5881748 0.8066746 1.0432544
# Plot the found matrix:
plot(g_map, type = "heatmap")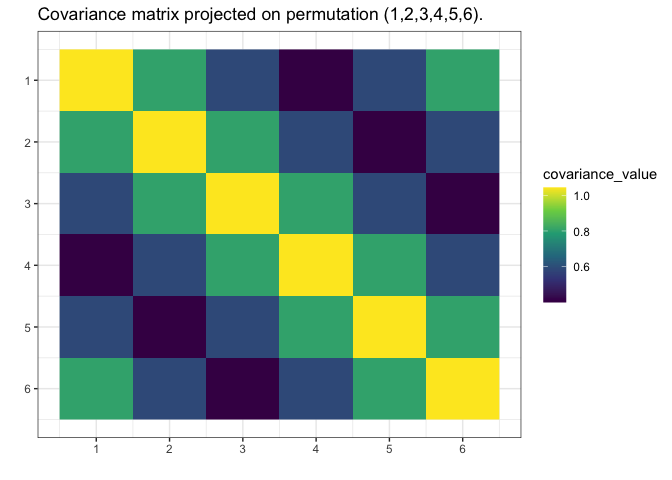
We see gips found the data’s structure, and we could
estimate the covariance matrix with huge accuracy only with a small
amount of data and additional reasonable assumptions.
Note that the rank of the S matrix was 4, while the rank
of the S_projected matrix was 6.
For more examples and introduction, see vignette("gips")
or its pkgdown
page.
Project was funded by Warsaw University of Technology within the Excellence Initiative: Research University (IDUB) programme.