Felix Cheysson
This package implements a spectral approach to the parametric estimation of Hawkes processes from Binned Observations through Whittle likelihood (HawkesBOW). It is based on the results of the article (Cheysson and Lang 2020).
You can install the released version of hawkesbow from GitHub with:
devtools::install_github("fcheysson/hawkesbow")Hawkes processes form a family of models for point processes for
which the occurrence of an event temporarily increases the probability
of future events occurring (Hawkes 1971). Formally, a Hawkes process
on
can be defined from its conditional intensity : where the variables denote the
arrival times of the process, the immigration intensity is a
positive constant, and the reproduction function
is a
measurable function. The reproduction function can be further decomposed
as
, where
is called the and
is a true density function,
, called the .
Alternatively, the Hawkes process can be constructed as a poissonian
cluster process (Hawkes and Oakes 1974). The process consists of a flow
of immigrants, the cluster centres, arriving according to a
homogeneous Poisson process of intensity . Then,
an immigrant arriving at time generates
children according to an inhomogeneous Poisson process of
intensity . These in turn independently generate children
according to the same process, and so on ad infinitum. The
processes consisting of an immigrant and all its descendants are
therefore branching processes, and are independent of each other.
Finally, the process resulting from the superposition of all these
branching processes is a Hawkes process of conditional intensity
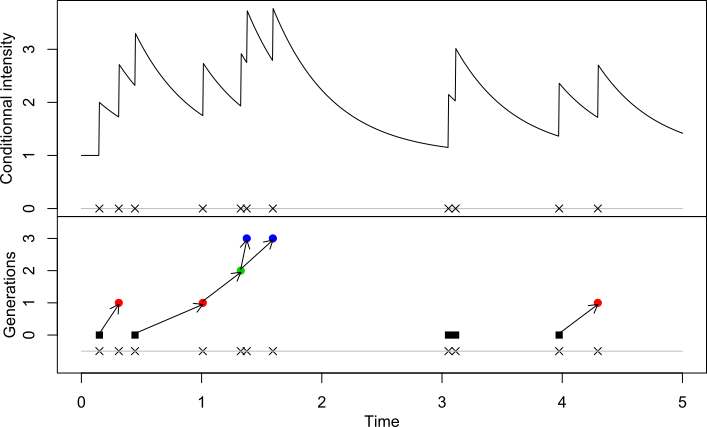
(see the following figure of a realisation of an
exponential Hawkes process with
, ).
This package also supports non-causal Hawkes processes, for which the
reproduction kernel
may take non-negative values on . Such processes are not defined through their
conditional intensity functions, but through the poisson cluster
representation, where each individual can potentially generate
offsprings both in the future and in the past.
The count sequence of a Hawkes process is the time series generated
by the event counts of the process, that is the series obtained by
counting the events of the process on intervals of fixed length.
Formally, the count sequence with bin size
associated to the point process is the
sequence .
This package fully supports the parametric estimation of Hawkes processes from their count sequences via minimisation of the Whittle likelihood, and partially supports the estimation from the arrival times via maximisation of the likelihood.
The parameters of a Hawkes process with arrival times on the interval can be estimated by maximising the following
pseudo-likelihood
Note that each step of the optimisation is usually of complexity .
Alternatively, the parameters of a Hawkes process with count sequence
can be estimated by minimising the
log-spectral (Whittle) likelihood
where
denotes the periodogram of
and
its spectral density function. Note that each
step of the optimisation is of complexity
and the periodogram can be calculated in
complexity
using a Fast Fourier Transform algorithm,
making this method usually faster than maximum likelihood
estimation.
This package supports both Hawkes processes and their count sequences.
Hawkes processes can be simulated by the function
hawkes:
hawkes(end, fun, repr, family, M=null, ...)where end denotes the end of the interval .
This function uses the cluster representation:
First, the immigrants are drawn according to a (potentially
inhomogeneous) Poisson process with intensity measure
fun.
Second, the number of offsprings of an immigrant are drawn from a
Poisson distribution with intensity repr.
Third, these offsprings are distributed according to the
family distribution.
Then, further offsprings are generated according to the last two steps.
The argument fun can take a numeric value or be
specified as a function, in which case the argument M must
be specified as an upper bound on fun (to allow simulation
of the immigration process by thinning). The argument
family can either be specified as a string
name corresponding to a distribution with random generation
function rname (for example exp for
rexp), or directly as a random generation function. The
optional arguments ... are passed to the random generation
function specified by family.
This returns a list of class hawkes, whose realisations
are stored in the member p. Other members of the list are
mainly used for support functions.
x <- hawkes(10, fun = 1, repr = 0.5, family = "exp", rate = 2)x <- hawkes(10, fun=function(y) {1+sin(y)}, M=2, repr=0.25,
family=function(n) {1 - sqrt(1 - runif(n))})plot.hawkes:plot.hawkes(x, intensity = FALSE, precision = 1e3, fun = NULL, repr = NULL, family = NULL,
M = NULL, ...)If x is of class hawkes, as for objects
returned by the function hawkes, arguments fun
through M can be ignored. If intensity is set
to FALSE, this plots the genealogy of the simulated Hawkes
process (as in preceding figure, bottom panel). If it is set to
TRUE, this plots the conditional intensity of the process
(as in preceding figure, top panel).
Two functions implement the estimation of Hawkes processes:
mle from arrival times
and whittle from count sequences .
While the optimisation procedure rely on existing functions (see below),
calculations of both the usual and Whittle likelihood functions are done
in C++ via Rcpp (Eddelbuettel and François 2011) and RcppArmadillo
(Eddelbuettel and Sanderson 2014).
The maximum likelihood method is implemented by the function
mle(events, kern, end, init = NULL, opts = NULL, ...)events holds the arrival times in
ascending order, kern must be a string (partially) matching
one of the reproduction kernels (see below) and end denotes
the endpoint of observation of the process.
The optimisation of the maximum likelihood function is done by the
function nloptr from the package nloptr (Johnson, n.d.)
with algorithm L-BFGS-B, where the derivatives of the likelihood are
calculated explicitly. By default, parameters are constrained to be
positive, and additionally is
constrained to be below 1. However, both the arguments opts
and ... are passed on to nloptr, so the
algorithm, the constraints, or any other parameter of the optimisation
procedure can be changed.
Example of use:
x = hawkes(100, fun = 1, repr = .5, family = "exp", rate = 1)
mle(x$p, "Exponential", x$end)The Whittle likelihood method is implemented by the function
whittle(counts, kern, binsize = NULL, trunc = 5L, init = NULL, ...)counts holds the count sequence ,
kern must be a string (partially) matching one of the
reproduction kernels (see below), binsize denotes the bin
size and trunc is the number of foldings due
to aliasing taken into account.
The optimisation of the Whittle likelihood function is done by the
function optim, with algorithm L-BFGS-B where the
derivatives of the likelihood are approximated by finite differences. By
default, parameters are constrained to be positive, and additionally
is constrained to be below 1. However, the argument ... is
passed to optim, so any optimisation parameter can be
changed.
Example of use:
x = hawkes(1000, fun = 1, repr = .5, family = "exp", rate = 1)
y = discrete(x, binsize = 1)
whittle(y, "Exponential", 1)Note that discrete is a useful function to create the
count sequence
associated with an object x of class
hawkes.
We introduce the reproduction kernels that are currently implemented
in this package. Recall that the Fourier transform of a reproduction
kernel is given by
and that it is a Hermitian function .
This is the exponential density function with parameter :
Its Fourier transform is
The exponential kernel can be specified with the string
Exponential and the parameter with the usual argument
rate. Both maximum
and Whittle likelihood methods are fully implemented for exponential
kernels. Moreover, the likelihood function is implemented in complexity
, using the relations in (Ozaki and Ogata
1979).
This is a symmetrised version of the exponential density function
with parameter :
Its Fourier transform is
The symmetric exponential kernel can be specified with the string
SymmetricExponential and the parameter with
the argument rate. Only the Whittle likelihood method is
implemented for symmetric exponential kernels. Note that it is a
non-causal kernel, as for .
This is the gaussian density function with mean and variance
:
Its Fourier transform is
The gaussian kernel can be specified with the string
Gaussian and its parameters and
with the usual arguments mean and
sd respectively. Only the Whittle likelihood method is
implemented for gaussian kernels. Note that it is a non-causal kernel,
as for .
This is a normalised and shifted power law function, with shape and scale :
For positive , its Fourier transform is given by
where denotes the integral
With successive integration by parts, this integral can be related to
, with
.
If or equivalently
, the integral , called the exponential integral with imaginary
argument, can be related the trigonometric integrals and calculated
using Padé approximants (Rowe et al. 2015, Appendix B), accurate to
better than .
The function
E1_imaginary implements this
approximation.
If or equivalently
, the integral
can be related to the incomplete gamma function
with imaginary argument
where . We implemented Taylor approximations of
this integral, accurate to better than ,
in the function
inc_gamma_imag.
The power law kernel can be specified with the string
PowerLaw and its parameters and with the arguments
shape and
scale respectively. Both maximum and Whittle likelihood
methods are implemented for power law kernels.
This is the Pareto density function with shape and scale :
For positive , its Fourier transform is given by
see above for the definition of .
Only Pareto kernels with fixed , , and have been implemented and can specified with the strings
Pareto1, Pareto2 and Pareto3
respectively, with parameter specified with
the argument scale. Only the Whittle method is available
for Pareto kernels.
Improve this vignette: it is currently too sparse and functions of the package could need some better description.
Add some real datasets to the package: real life case-studies with good datasets help understand the functionalities of a package.
Variance and confidence intervals for the estimation with
function whittle: note that currently, the
variance-covariance matrix returned by the optimisation method in
function whittle is not accurate, as it ignores the
dependence within the count sequence .
Diagnostics for the estimated model: spectral density based goodness-of-fit tests are to be implemented for the estimated Hawkes processes, based on the work of (Paparoditis 2000).
Custom built-kernels: allow the user to input reproduction kernels that are not already implemented.
Cheysson, Felix, and Gabriel Lang. 2020. “Strong mixing condition for Hawkes processes and application to Whittle estimation from count data.” arXiv, March. https://arxiv.org/abs/2003.04314.
Eddelbuettel, Dirk, and Romain François. 2011. “Rcpp : Seamless R and C++ Integration.” J. Stat. Softw. 40 (8): 1–18. https://doi.org/10.18637/jss.v040.i08.
Eddelbuettel, Dirk, and Conrad Sanderson. 2014. “RcppArmadillo: Accelerating R with High-Performance C++ Linear Algebra.” Computational Statistics and Data Analysis 71: 1054–63. http://dx.doi.org/10.1016/j.csda.2013.02.005.
Hawkes, Alan G. 1971. “Spectra of Some Self-Exciting and Mutually Exciting Point Processes.” Biometrika 58 (1): 83–90. https://doi.org/10.2307/2334319.
Hawkes, Alan G., and David Oakes. 1974. “A cluster process representation of a self-exciting process.” J. Appl. Probab. 11 (03): 493–503. https://doi.org/10.2307/3212693.
Johnson, Steven G. n.d. “The NLopt nonlinear-optimization package.”
Ozaki, T., and Yosihiko Ogata. 1979. “Maximum likelihood estimation of Hawkes’ self-exciting point processes.” Ann. Inst. Stat. Math. 31 (1): 145–55. https://doi.org/10.1007/BF02480272.
Paparoditis, Efstathios. 2000. “Spectral density based goodness-of-fit tests for time series models.” Scand. J. Stat. 27 (1): 143–76. https://doi.org/10.1111/1467-9469.00184.
Rowe, B. T. P., M. Jarvis, R. Mandelbaum, G. M. Bernstein, J. Bosch, M. Simet, J. E. Meyers, et al. 2015. “GalSim: The modular galaxy image simulation toolkit.” Astron. Comput. 10: 121–50. https://doi.org/10.1016/j.ascom.2015.02.002.