The goal of segregatr is to provide segregation analysis for clinical variant classification.
You can install segregatr from CRAN as follows:
install.packages("segregatr")Alternatively, you can obtain the latest development version from GitHub:
devtools::install_github("magnusdv/segregatr")We start by loading segregatr:
library(segregatr)
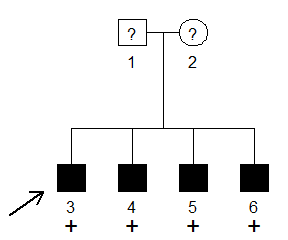
#> Loading required package: pedtoolsThe family below shows four brothers, all affected with a rare dominant disease with 90% penetrance and phenocopy rate 1%. The parents have unknown affection status. All four brothers are shown to carry a candidate variant, warranting a segregation analysis. pathogenic variant.

In order to compute the full-likelihood Bayes factor, we first create the pedigree.
x = nuclearPed(4)Then we run the FLB() function, filling in the necessary
data:
FLB(x, carriers = 3:6, aff = 3:6, unknown = 1:2,
freq = 0.0001, penetrances = c(0.01, 0.9, 0.9), proband = 3)
#> [1] 7.732161The answer indicates only suggestive evidence for pathogenicity.