



The x3p file format is specified in ISO standard 5436:2000 to describe 3d surface measurements. This package allows reading, writing and basic modifications to the 3D surface measurements.
x3ptools is available from CRAN:
install.packages("x3ptools")The development version is available from Github:
# install.packages("devtools")
devtools::install_github("heike/x3ptools", build_vignettes = TRUE)The x3p file format is an xml based file format created to describe digital surface measurements. x3p has been developed by OpenFMC (Open Forensic Metrology Consortium, see https://www.open-fmc.org/) and has been adopted as ISO ISO5436 – 2000. x3p files are a zip archive of a directory consisting of an xml file of meta information and a matrix of numeric surface measurements.
Internally, x3p objects are stored as a list consisting of the surface matrix (the measurements) and meta information in four records: header info, feature info, general info, and matrix info:
library(x3ptools)
logo <- read_x3p(system.file("csafe-logo.x3p", package="x3ptools"))
names(logo)## [1] "header.info" "surface.matrix" "feature.info" "general.info"
## [5] "matrix.info"The four info objects specify the information for Record1 through
Record4 in the xml file. An example for an xml file is provided with the
package and can be accessed as
system.file("templateXML.xml", package="x3ptools").
header.info contains the information relevant to
interpret locations for the surface matrix:
logo$header.info## $sizeY
## [1] 419
##
## $sizeX
## [1] 741
##
## $incrementY
## [1] 6.45e-07
##
## $incrementX
## [1] 6.45e-07matrix.info expands on header.info and
provides the link to the surface measurements in binary format.
general.info consists of information on how the data was
captured, i.e. both author and capturing device are specified here.
feature.info is informed by the header info and provides
the structure for storing the information.
While these pieces can be changed and adapted manually, it is more
convenient to save information on the capturing device and the creator
in a separate template and bind measurements and meta information
together in the command addtemplate_x3p.
read_x3p and write_x3p are the two
functions allows us to read x3p files and write to x3p files.
logo <- read_x3p(system.file("csafe-logo.x3p", package="x3ptools"))
names(logo)## [1] "header.info" "surface.matrix" "feature.info" "general.info"
## [5] "matrix.info"The function image_x3p uses rgl to render a
3d object in a separate window. The user can then interact with the 3d
surface (zoom and rotate):
image_x3p(logo, size=c(741,419), zoom=0.5, useNULL=TRUE)
rglwidget()
In case a file name is specified in the function call the resulting surface is saved in a file (the extension determines the actual file format of the image).
image_x3p_grid lays a regularly spaced grid of lines
over the surface of the scan. Lines are drawn spaces apart
(50 microns by default in y direction and 100 microns in x direction).
Every fifth and tenth lines are colored differently to ease a visual
assessment of distance.
logoplus <- x3p_add_grid(logo, spaces=50e-6,
size = c(3,3,5), color=c("grey50", "black", "darkred"))
image_x3p(logoplus, size=c(741,419), zoom=0.5, useNULL=TRUE)
rglwidget()
The functions x3p_to_df and df_to_x3p allow
casting between an x3p format and an x-y-z data set:
logo_df <- x3p_to_df(logo)
head(logo_df)## x y value
## 1 0.000e+00 0.00026961 4e-07
## 2 6.450e-07 0.00026961 4e-07
## 3 1.290e-06 0.00026961 4e-07
## 4 1.935e-06 0.00026961 4e-07
## 5 2.580e-06 0.00026961 4e-07
## 6 3.225e-06 0.00026961 4e-07When converting from the x3p format to a data frame, the values from
the surface matrix are interpreted as heights (saved as
value) on an x-y grid. The dimension of the matrix sets the
number of different x and y levels, the information in
header.info allows us to scale the levels to the measured
quantities. Similarly, when moving from a data frame to a surface
matrix, the assumption is that measurements are taken on an equi-spaced
and complete grid of x-y values. The information on the resolution
(i.e. the spacing between consecutive x and y locations) is saved in
form of the header info, which is added to the list. General info and
feature info can not be extracted from the measurements, but have to be
recorded with other means.
Once data is in a regular data frame, we can use our regular means to
visualize these raster images, e.g. using ggplot2:
library(ggplot2)
library(magrittr)
logo_df %>% ggplot(aes( x= x, y=y, fill= value)) +
geom_tile() +
scale_fill_gradient2(midpoint=4e-7)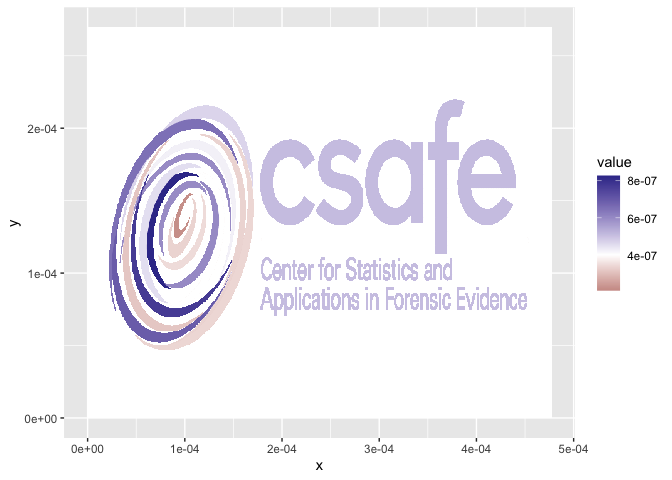
rotate_x3p rotates an x3p image in steps of 90 degrees,
transpose_x3p transposes the surface matrix of an image and
updates the corresponding meta information. The function
y_flip_x3p is a combination of transpose and rotation, that
allows to flip the direction of the y axis to move easily from legacy
ISO x3p scans to ones conforming to the most recent ISO standard.
sample_x3p allows to sub-sample an x3p object to get a
lower resolution image. In sample_x3p we need to set a
sampling factor. A sample factor mX of 2 means that we only
use every 2nd value of the surface matrix, mX of 5 means,
we only use every fifth value:
dim(logo$surface.matrix)## [1] 741 419logo_sample <- sample_x3p(logo, m=5)
dim(logo_sample$surface.matrix)## [1] 149 84Parameter mY can be specified separately from
mX for a different sampling rate in y direction. We can
also use different offsets in x and y direction by specifying
offsetX and/or offsetY.
sample2 <- sample_x3p(logo, m=5, offset=1)
cor(as.vector(logo_sample$surface.matrix[-149,]), as.vector(sample2$surface.matrix))## [1] 0.8639308interpolate_x3p allows, like sample_x3p, to
create a new x3p file at a new resolution, as specified in the
parameters resx and resy. The new resolution
should be lower (i.e. larger values for resx and
resy) than the resolution specified as
IncrementX and IncrementY in the header info
of the x3p file. interpolate_x3p can also be used to
interpolate missing values (set parameter maxgap according
to specifications in zoo::na.approx).
The four records of header info, feature info, general info, and
matrix info of an x3p object contain meta information relevant to the
scan. x3p_show_xml allows the user to specify a search term
and then proceeds to go through all records for a matching term. As a
result any matching elements from the meta file are shown:
logo %>% x3p_show_xml("axis") # three axes are defined## $Axes.CX.AxisType
## [1] "I"
##
## $Axes.CY.AxisType
## [1] "I"
##
## $Axes.CZ.AxisType
## [1] "A"logo %>% x3p_show_xml("creator")## $Creator
## [1] "Heike Hofmann, CSAFE"If a search term identifies a single element,
x3p_modify_xml allows the user to update the corresponding
meta information:
logo %>% x3p_modify_xml("creator", "I did this")## x3p object
## Instrument: N/A
## size (width x height): 741 x 419 in pixel
## resolution: 6.4500e-07 x 6.4500e-07
## Creator: I did this
## Comment: image rendered from the CSAFE logoA mask is used to markup regions of the surface - the mask can then be used to overlay the rendered surface with color.
Below is a 3d surface of a part of a bullet surface is shown. Some regions are marked up: the bronze colored area is an area with strong striae (vertical lines engraved on the bullet as it moves through the barrel of the handgun when fired), areas in dark blue show groove engraved areas, the light blue area shows break off at the bottom of the bullet, and the pink area marks an area without striae:

Any image can serve as a mask, the command x3p_add_mask
allows to add a raster image as a mask for a x3p object:
logo <- read_x3p(system.file("csafe-logo.x3p", package="x3ptools"))
color_logo <- png::readPNG(system.file("csafe-color.png", package="x3ptools"))
logoplus <- x3p_add_mask(logo, mask = as.raster(color_logo))
image_x3p(logoplus, size=c(741, 419), zoom=0.5, multiply = 30)
Some masks are more informative than others, but the only requirement for images is that they are of the right size.


Masks are raster images. Any change to the raster manifests as a
change in the surface color of the corresponding scan. We suggest using
the magick package to manipulate masks.
Additionally, vertical and horizontal lines can be added in the masks
using the commands x3p_add_vline and
x3p_add_hline:
logo <- read_x3p(system.file("csafe-logo.x3p", package="x3ptools"))
logoplus <- x3p_add_hline(logo, yintercept=c(13e-5,19.5e-5), color="cyan")
#image_x3p(logoplus, size=c(741, 419)/2, zoom=0.5, multiply = 30, file="man/figures/logo-lines.png")