The goal of ravetools is to provide memory-efficient
signal processing toolbox for intracranial-EEG analyses.
Highlighted features include:
Notch filter (remove electrical line
frequencies)Welch Periodogram (averaged power over
frequencies)Wavelet (frequency-time
decomposition)The package is available on CRAN. To install the
compiled version, simply run:
install.packages("ravetools")Installing the package from source requires installation of proper
compilers and some C libraries; see this
document for details.
iEEG preprocess
pipelineThis is a basic example which shows you how to preprocess an
iEEG signal. The goal here is to:
* Channel referencing is not included
library(ravetools)
# Generate 20 second data at 2000 Hz
time <- seq(0, 20, by = 1 / 2000)
signal <- sin( 120 * pi * time) +
sin(time * 20*pi) +
exp(-time^2) *
cos(time * 10*pi) +
rnorm(length(time))
diagnose_channel(signal, srate = 2000)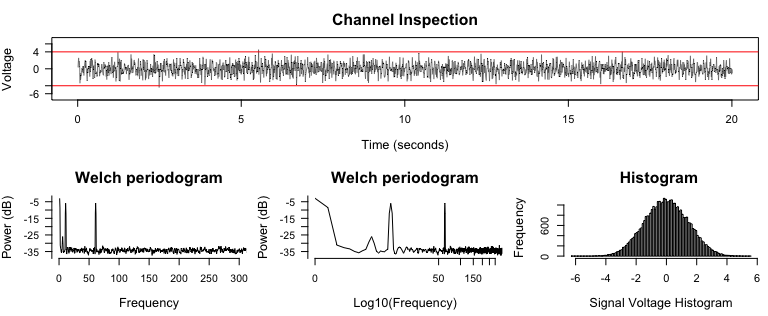
Notch filters and inspect Periodograms## ------- Notch filter --------
signal2 <- notch_filter(signal, sample_rate = 2000)
diagnose_channel(signal, signal2, srate = 2000,
name = c("Raw", "Filtered"))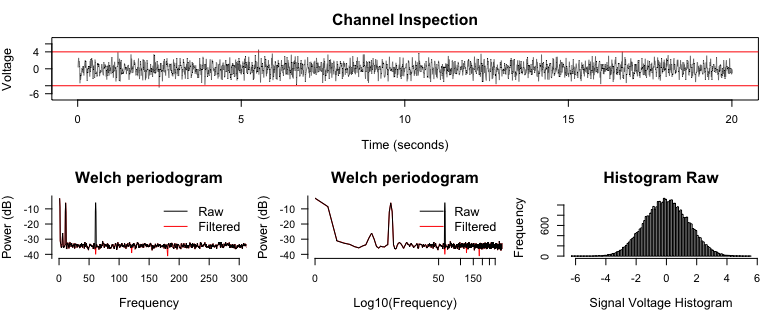
Current version of ravetools provides two approaches:
Wavelet and Multi-taper. Wavelet
uses the Morlet wavelet
and obtains both amplitude and phase data, while
Multi-taper does not generate phase data. However, the
amplitude obtained from Multi-taper is smoother than
Wavelet.
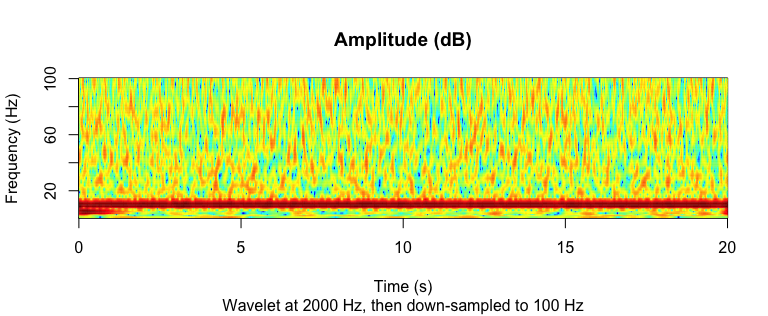
Wavelet:## ---------- Wavelet -----------
coef <- morlet_wavelet(
signal2, freqs = seq(1, 100, by = 1),
srate = 2000, wave_num = c(2, 15))
amplitude <- 20 * log10(Mod(coef[]))
# For each frequency, decimate to 100 Hz
downsample_amp <- apply(amplitude, 2, decimate, q = 20)
downsample_time <- decimate(time, q = 20)
par(mfrow = c(1,1))
image(
z = downsample_amp,
x = downsample_time,
y = seq(1, 100, by = 1),
xlab = "Time (s)",
ylab = "Frequency (Hz)",
main = "Amplitude (dB)",
sub = "Wavelet at 2000 Hz, then down-sampled to 100 Hz",
col = matlab_palette()
)
Multi-taperAlternatively you can use Multi-tapers to obtain
amplitude data. The algorithm is modified from source code here. Please
credit them as well if you adopt this approach.
## ---------- Multitaper -----------
res <- multitaper(
data = signal2,
fs = 2000,
frequency_range = c(1, 100),
time_bandwidth = 1.5,
window_params = c(2, 0.01),
nfft = 100
)
par(mfrow = c(1,1))
image(
x = res$time,
y = res$frequency,
z = 10 * log10(res$spec),
xlab = "Time (s)",
ylab = 'Frequency (Hz)',
col = matlab_palette(),
main = "Amplitude (dB)"
)
RAVE
paper from Beauchamp's labMagnotti, JF, and Wang, Z, and Beauchamp, MS. RAVE: comprehensive
open-source software for reproducible analysis and visualization of
intracranial EEG data. NeuroImage, 223, p.117341.The multitaper function uses the script derived from
Prerau's lab. The TinyParallel script is
derived from RcppParallel package with TBB
features removed (only use tinythreads).
[1] Magnotti, JF, and Wang, Z, and Beauchamp, MS. RAVE: comprehensive
open-source software for reproducible analysis and visualization of
intracranial EEG data. NeuroImage, 223, p.117341.
[2] Prerau, Michael J, and Brown, Ritchie E, and Bianchi, Matt T, and
Ellenbogen, Jeffrey M, and Purdon, Patrick L. Sleep Neurophysiological
Dynamics Through the Lens of Multitaper Spectral Analysis. Physiology,
December 7, 2016, 60-92.
[3] JJ Allaire, Romain Francois, Kevin Ushey, Gregory Vandenbrouck, Marcus
Geelnard and Intel (2022). RcppParallel: Parallel Programming Tools for
'Rcpp'. R package version 5.1.5.
https://CRAN.R-project.org/package=RcppParallel